Software Quantifies Leaf Vein Networks, Enables Plant Biology Advances
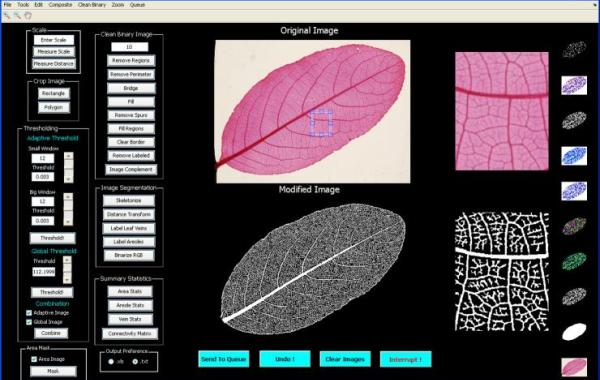
LEAF GUI screenshot
Plant biologists are facing pressure to quantify the response of plants to changing environments and to breed plants that can respond to such changes. One method of monitoring the response of plants to different environments is by studying their vein network patterns. These networks impact whole plant photosynthesis and the mechanical properties of leaves, and vary between species that have evolved or have been bred under different environmental conditions.
To help address the challenge of how to quickly examine a large quantity of leaves, researchers at the Georgia Institute of Technology have developed a user-assisted software tool that extracts information about macroscopic vein structures directly from leaf images.
"The software can be used to help identify genes responsible for key leaf venation network traits and to test ecological and evolutionary hypotheses regarding the structure and function of leaf venation networks," said Joshua Weitz, an assistant professor in the Georgia Tech School of Biology.
The program, called Leaf Extraction and Analysis Framework Graphical User Interface (LEAF GUI), enables scientists and breeders to measure the properties of thousands of veins much more quickly than manual image analysis tools.
Details of the LEAF GUI software program were published in the "Breakthrough Technologies" section of the January issue of the journal Plant Physiology. Development of the software, which is available for download at www.leafgui.org, was supported by the Defense Advanced Research Projects Agency (DARPA) and the Burroughs Welcome Fund.
LEAF GUI is a user-assisted software tool that takes an image of a leaf and, following a series of interactive steps to clean up the image, returns information on the structure of that leaf's vein networks. Structural measurements include the dimensions, position and connectivity of all network veins, and the dimensions, shape and position of all non-vein areas, called areoles.
"The network extraction algorithms in LEAF GUI enable users with no technical expertise in image analysis to quantify the geometry of entire leaf networks -- overcoming what was previously a difficult task due to the size and complexity of leaf venation patterns," said the paper's lead author Charles Price, who worked on the project as a postdoctoral fellow at Georgia Tech. Price is now an assistant professor of plant biology at the University of Western Australia.
While the Georgia Tech research team is currently using the software to extract network and areole information from leaves imaged under a wide range of conditions, LEAF GUI could also be used for other purposes, such as leaf classification and description.
"Because the software and the underlying code are freely available, other investigators have the option of modifying methods as necessary to answer specific questions or improve upon current approaches," said Price.
LEAF GUI is not the only software program Weitz's group has developed to investigate the network characteristics of plants. In March 2010, Weitz's group co-authored another "Breakthrough Technologies" paper in Plant Physiology detailing a way to analyze the complex root network structure of crop plants, with a focus on rice.
This work was performed in collaboration with Anjali Iyer-Pascuzzi, John Harer and Philip Benfey at Duke University and was supported by DARPA, the National Science Foundation and the Burroughs Welcome Fund.
"Both of these software programs are enabling tools in the growing field of 'plant phenomics,' which aims to correlate gene function, plant performance and response to the environment," noted Weitz. "By identifying leaf vein characteristics and root structures that differ between plants, we are enabling advances in basic plant science and, in the case of crop plants, assisting researchers in identifying and potentially altering genes to improve plant health, yield and survival."
In addition to those already mentioned, Olga Symonova, Yuriy Mileyko and Troy Hilley also contributed to this work at Georgia Tech.
These projects were supported by the Defense Advanced Research Projects Agency (DARPA) (Award No. HR0011-05-1-0057), National Science Foundation (NSF Plant Genome Research Program Award Nos. 0606873 and 0820624) and Burroughs Wellcome Fund (BWF). The content is solely the responsibility of the principal investigator and does not necessarily represent the official views of DARPA, NSF or BWF.
Research News & Publications Office
Georgia Institute of Technology
75 Fifth Street, N.W., Suite 314
Atlanta, Georgia 30308 USA
Media Relations Contacts: Abby Robinson (abby@innovate.gatech.edu; 404-385-3364) or John Toon (jtoon@gatech.edu; 404-894-6986)
Writer: Abby Robinson
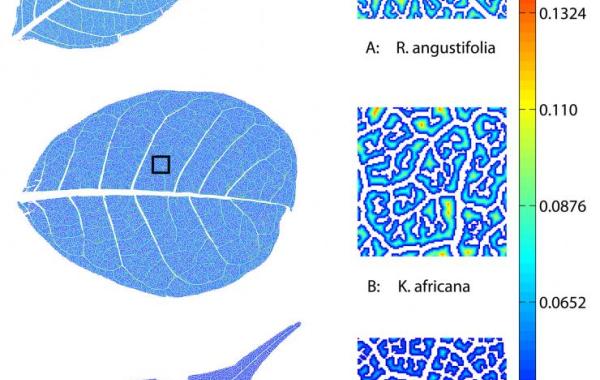
LEAF GUI example
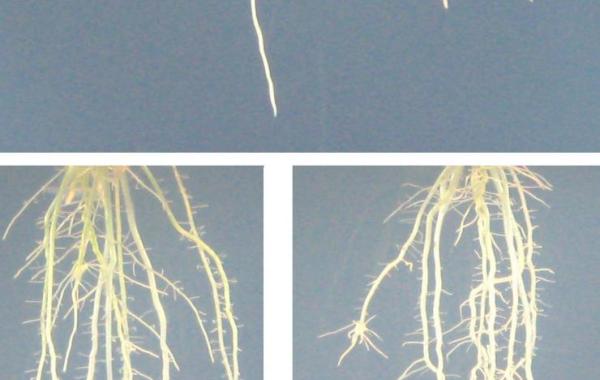
rice root system architecture
